During my research internship at the Harvard SEAS Visual Computing Group, I had the opportunity to work on an exciting project called Visual Analytics for Tree Comparison. The project was aimed at creating a visual analytics tool for semi-automated tree comparison with a specific use case for in vitro fertilization datasets.
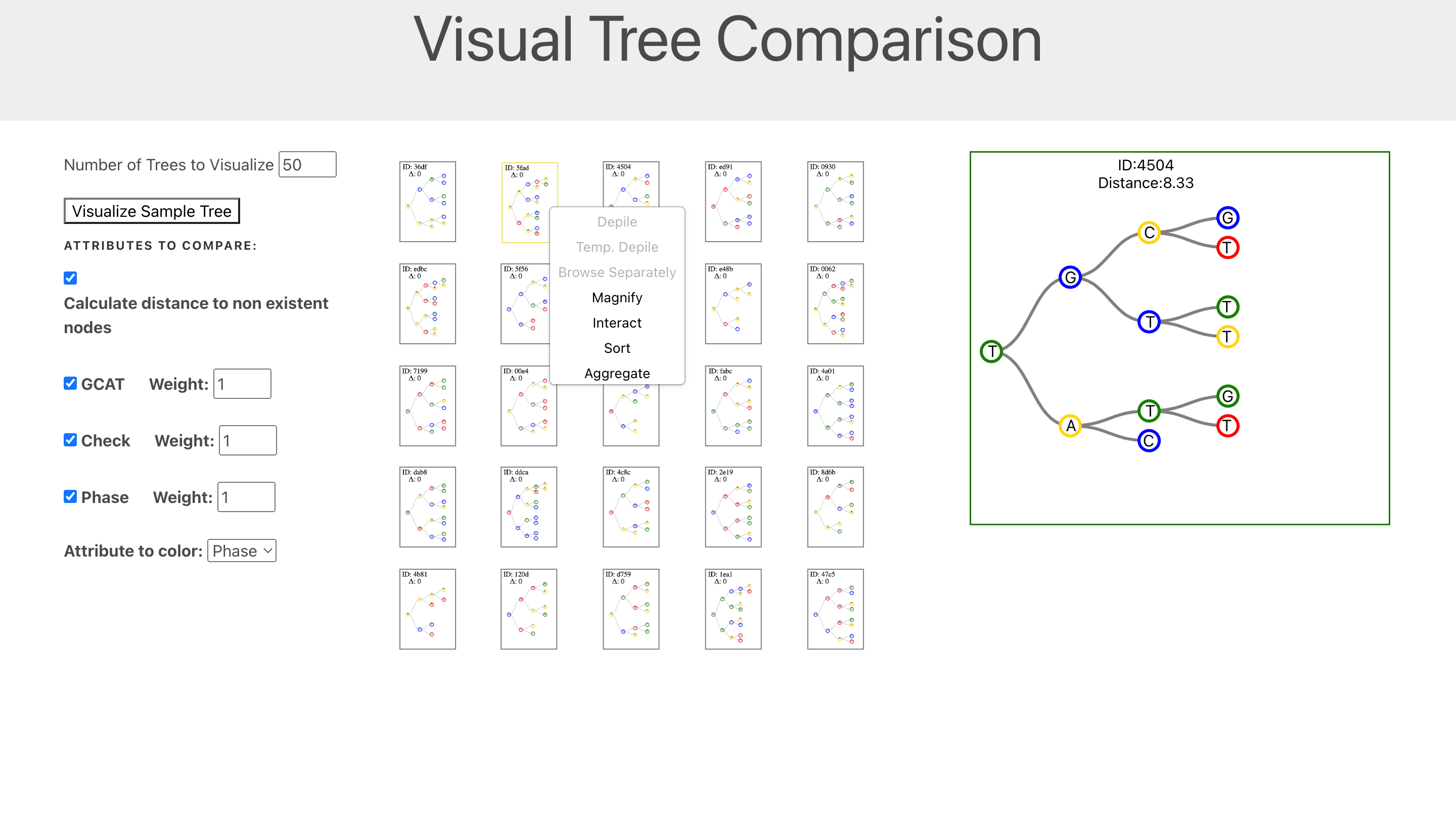 As you can see above, the tool allows users to compare multiple trees and visualize the differences between them. The tool also allows users to cluster trees based on their similarity. A difficulty I faced while developing this project was the lack of available datasets that I could use to test my visualizations. This was because of the privacy laws concering medical data such as IVF datasets. To overcome this issue, I created a synthetic dataset generator that could generate random trees with random cell data. This allowed me to test my visualizations and ensure that they were working as intended.
As you can see above, the tool allows users to compare multiple trees and visualize the differences between them. The tool also allows users to cluster trees based on their similarity. A difficulty I faced while developing this project was the lack of available datasets that I could use to test my visualizations. This was because of the privacy laws concering medical data such as IVF datasets. To overcome this issue, I created a synthetic dataset generator that could generate random trees with random cell data. This allowed me to test my visualizations and ensure that they were working as intended.
As the full-stack developer for the project, I built a Python Flask backend and a Node.js JavaScript frontend. I also conducted research on binary tree visualization and comparison for medical applications, and implemented a Zhang-Shasha edit distance metric to optimize tree clustering tools.
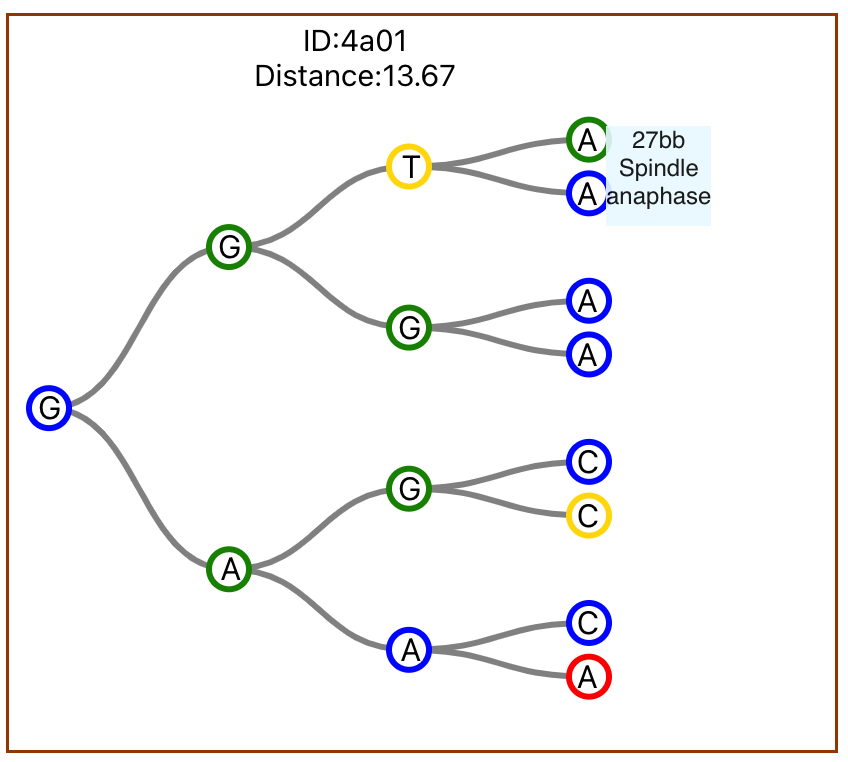 Additionally, users of the platform can also interact with specific trees to view more information about the cells in the tree. They can hover over specified nodes to view information and can also resort the set of trees based on this cell data.
Additionally, users of the platform can also interact with specific trees to view more information about the cells in the tree. They can hover over specified nodes to view information and can also resort the set of trees based on this cell data.
To enable clustering for the compared trees, we worked with piling.js. Additionally, we leveraged d3.js visualization and Python libraries to perform complex medical computations for cell data comparison.
Throughout the project, we also collaborated with systems pharmacologists at Harvard LSP, further enhancing the scope and impact of our work. Overall, Visual Analytics for Tree Comparison was a fascinating project to work on and provided valuable experience in developing tools for medical research.
This project’s code is private, but you can find the project website here.